Sophia Cottrill
Age 15 | Tara, Ontario
Canada-Wide Science Fair Intermediate Excellence Award: Silver Medallist
Biofluorescence in marine fish is a recently discovered phenomenon (Sparks et al., 2014), but has not been studied in freshwater fish. In this study, 128 specimens of freshwater biota from 12 locations around Lake Huron and Georgian Bay were photographed under ultraviolet light. It was discovered that biofluorescence is widespread across freshwater taxonomic families and species. The discovery of these fluorescent pigments is important to developing biomedical imaging technology.
BACKGROUND
Biofluorescence in marine fish is a recently discovered phenomenon (Sparks et al., 2014), but it has yet to be studied in freshwater species of fish. Biofluorescence is the absorption of light, and the re-emission of this light at a longer wavelength of lower energy by a living organism (Sparks et al., 2014). The purpose of this project was to determine whether freshwater species from Lake Huron and Georgian Bay exhibit fluorescence under ultraviolet (UV) light, similar to marine species of fish in the Cayman Islands (Sparks et al., 2014). This fluorescence is due to the presence of fluorophores in organisms (Tsien, 1998), and can be correlated to the phylogenetic relationships and intraspecific communication between species (Sparks et al., 2014). This project aims to identify the genotypes and phenotypes associated with fluorescence in freshwater species. More specifically, focus was placed upon patterns and anatomical locations of fluorescence, natural behaviours and adaptations, and phylogenetic relationships as components for investigating fluorescence in Great Lakes biota. Biofluorescence has numerous biomedical applications. Certain fluorescent proteins such as green fluorescent proteins, have specific properties such as emission wavelengths or environmental conditions that make them valuable to modern day medicine for tagging, imaging, and quantifying intracellular protein (i.e. transmembrane protein channels) and organelle (i.e. mitochondria) activity (Tsien, 1998). Studying fluorescence in biota is beneficial to furthering medical technology and understanding the hidden function of biofluorescence in the natural environment.
HYPOTHESIS
I hypothesized that i) freshwater species of fish would exhibit fluorescence when photographed under UV light, ii) closely related species that look alike under ambient light would have distinguishing fluorescent patterns (Siebeck et al., 2010), iii) the complexity, locations, patterns, and prominence of fluorescence would vary amongst relevant freshwater specimens.
METHODS
Part 1: Assessment of Biofluorescence
One hundred and twenty-eight specimens of twenty-six species of freshwater fish were photographed under both natural and UV light. These specimens were collected from twelve locations sampled in surveys conducted by the Ontario Ministry of Natural Resources and Forestry, the Nottawasaga Valley Conservation Authority, and the U.S. Geological Survey. A Nikon D3300 Digital SLR Camera equipped with a Yashica CS-240 auto electronic flash was used to photograph the fish specimens. The flash was equipped with a Hoya U-340 + Schott S8612 UV-Only Flash Stack filter that blocked all light except light with wavelengths between 300 and 400 nm (UV light). The camera was adapted for UV photography using the Schott BG38 52mm x 2mm UV/IR-Cut Filter Visual Bandpass IR Suppress filter to eliminate all UV light between 300 and 400 nm from the picture so that only visible light between 400 and 700 nm was recorded in the image. This camera system operated as part of a portable photo lab (Dimensions: 81.3cm x 45.7cm x 63.5cm) that was built from wood to photograph fish at a variety of different locations and ensure that images were captured in complete darkness. Computational software was used to remotely operate the camera to acquire accurate, focused photos. Fluorescence patterns of each specimen were quantified by determining if fluorescence was evident at 18 predetermined anatomical locations. The phenotypes of fluorescence in these species were then characterized by calculating the average number of fluorescent areas by taxonomic family and by individual species, allowing for comparison with the genetic analysis.
Part 2: Phylogenetic Analyses
The phylogenetic analyses closely followed those described by Sparks et al. (2014). The analyses were completed using sequences and tools available from GenBank®. Unfortunately, several of the genes identified by Sparks et al. (2014) were unavailable for the freshwater species sampled as part of this study. Analyses were conducted for cytochrome c oxidase 1 (COX1), which was associated with the International Barcode of Life Project (Hebert et al., 2003). As such, these particular nucleotide and protein sequences were well documented for many species of fish and invertebrates. The Basic Local Alignment Search Tool (BLAST®) was used to compare the sequence of the base pairs or amino acids to the sequences of the same nucleotide or protein for other species in the database for each of the twenty-six species. This allowed the similarity and relative proximity of two or more species to be measured at the molecular level for the gene in question.
RESULTS
All twenty-six species of freshwater biota photographed in this study were found to exhibit some degree of fluorescence when photographed under UV light, and there were many consistencies among a number of the species. The most common locations of fluorescence included the first dorsal fin, the caudal fin, and the operculum. Seventy-three percent of all specimens exhibited fluorescence on their first dorsal fin and on the caudal fin. Seventy-two percent of all specimens (n=92) exhibited fluorescence on their operculum (also known as the gill cover, located behind the eye on each side of the head).
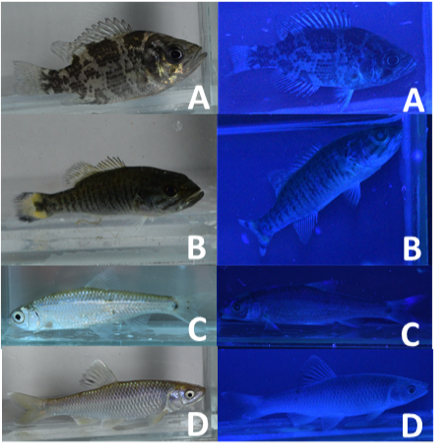
When summarized by taxonomic family (see Figure 1.), Salmonidae had the highest average number of areas of fluorescence at 13.0 ± 1.00 (n=3). Osmeridae also had a high average number of areas of fluorescence at 6.8 ± 5.36 (n=5). Fundulidae had the lowest average number of areas of fluorescence among vertebrate families in this study, averaging just 2.5 ± 0.71 (n=2). Centrarchidae species were found to average 5.9 ± 2.63 (n=48) areas of fluorescence. Within this family, Smallmouth Bass (Micropterus dolomieu) had an average of 8.0 ± 2.00 (n=9) areas of fluorescence, and Rock Bass (Ambloplites rupestris) had an average of 7.1 ± 2.88 (n=10). Cyprinidae species averaged 6.4 ± 2.62 (n=54) areas of fluorescence, with the highest species-specific average for Spottail Shiner (Notropis hudsonius) at 8.6 ± 1.14 (n=5), and Spotfin Shiner (Cyprinella spiloptera) at 7.9 ± 3.33 (n=9) on average. These species had a notably higher average number of areas of fluorescence than other members of the minnow family.
Figure 1. Average number of fluorescent areas among freshwater biota characterized by Taxonomic Family.
Figure 2. Fluorescent pigmentation in Great Lakes fish specimens. A, Rock Bass (Ambloplites rupestris); B, Smallmouth Bass (Micropterus dolomieu); C, Spottail Shiner (Notropis hudsonius); D, Spotfin Shiner (Cyprinella spiloptera).
DISCUSSION
This study aimed to identify the genotypes and phenotypes associated with biofluorescence in freshwater species of fish in Lake Huron and Georgian Bay. Biofluorescence was first studied in the Cayman Islands (Sparks et al., 2014), but had not been studied in freshwater species prior to this study, perhaps because the pigmentation of freshwater biota under ambient light is rather mundane compared to marine species. In this study, it was found that all vertebrate families averaged between 3.0 and 6.4 areas of fluorescence. The species that fluoresced the most among these taxonomic families belonged to the Cyprinidae and Centrarchidae families. The results of the phylogenetic analyses completed using GenBank® and BLAST® provided insight into the molecular identities of the freshwater species included in this study. The cladogram (Figure 3) demonstrates how closely related two species are by the cumulative distance from one another. This data can then be used to compare the fluorescence emission of species and observe patterns or intricate differentiation between closely related species. Generally, species that were shown to be similar at the molecular level exhibited comparable fluorescent pigmentation under UV light. These similarities in fluorescent emissions compared very closely with similarities in molecular composition among these species, as Centrarchidae species were found to be closely related at the molecular level and have similar fluorescent structures. Similarly, this idea applied to Cyprinidae species as well. These species were highly similar at the molecular level and also had comparable fluorescent pigmentation.
Figure 3. Phylogenetic analysis of freshwater species analysed in this study.
In this study, new sources of fluorescent pigments have been identified in twenty-six species of freshwater biota. It is probable that these fluorescent pigments will have various different genetic and physical properties as suggested by differences in pigmentation observed among taxon, species, and individual specimens. As these new sources and fluorescence types are explored, biomedical technologies continue to become strengthened and more diversified. Research to study functions within a cell (in vivo), has been conducted by using fluorescent proteins that are isolated from natural sources such as fish or corals. More specifically, experiments have been done using green fluorescent proteins (GFP) to study the way cancer spreads by imaging malignant cells moving through blood vessels (Hoffman, 2008, 2015; Cohen, 2016). Moreover, fluorescent proteins have been used to investigate how viruses such as HIV function so that potential cures can be assessed (Cambell et al., 2006; Cohen, 2016). Select properties of fluorescent proteins, such as those of the red fluorescent protein (RFP), are rare but valuable to medicine for applications such as illuminating neurons in the human brain (Miyawaki, 2005 & Cohen, 2016). Fluorescent proteins can be used to image activity and signalling among various cells and tissues by connecting fluorescent proteins to functional proteins to study their functions and behaviours in the human body (Miyawaki, 2005). By studying fluorescent properties of freshwater species, novel sources of fluorescent proteins could be identified, allowing the diverse array of fluorescent-based technologies to become more accessible and applicable in contemporary medical research and practice.
CONCLUSIONS
As I hypothesized, biofluorescence was found in freshwater biota with a variety of phenotypes and genotypes among twenty-six species and 128 specimens. Freshwater species of fish clearly exhibited fluorescence under UV light, as initially predicted. This fluorescence revealed some patterns amongst species and taxonomic groups. I found that species that look alike under ambient light and are closely related often had similar fluorescent pigmentation. Different intensities and locations of fluorescence were found to be comparable to marine biota (Sparks et al., 2014) as hypothesized, but all fluorescence exhibited was blue pigmentation. No green or red fluorescence was observed in this study. Fluorescence is thought to serve a variety of purposes in the natural underwater marine environment, including camouflage and mating (Sparks et al., 2014), lures (Haddock & Dunn, 2015), signalling and communication (Michiels et al., 2008), and species differentiation using ultraviolet light to discriminate between species that look very similar under natural light (Siebeck et al., 2010). It is probable that these natural processes of intraspecific communication could also exist in freshwater ecosystems.
ACKNOWLEDGEMENTS
Thank you to the Ministry of Natural Resources and Forestry, the Nottawasaga Valley Conservation Authority, the United States Geological Survey, Laurissa Christie, Adam, Anji, and Maisie Cottrill, Brad and Patti-Jo Lacey, and John Twelves for their assistance with this project.
REFERENCES
Cambell, E.M., Perez, O., Melar, M., & Hope, T.J. (2006). Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Northwestern University, Chicago, IL: Feinberg School of Medicine.
Cohen, C. (2016). Creatures of Light. NOVA: American Museum of Natural History.
Haddock, S. H. D. & Dunn, C. W. (2015). Fluorescent proteins function as a prey attractant: experimental evidence from the hydromedusa Olindias formosus and other marine organisms. The Company of Biologists.
Hebert, P. N. D., Ratnasingham, S., & deWaard, J. R. (2003). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. DNA divergences, 270, 96 – 99.
Hoffman, R. M. (2008). Imaging In Mice With Fluorescent Proteins: From Macro To Subcellular. University of California, San Diego: AntiCancer, Inc., Surgery Dept.
Hoffman, R. M. (2015). Application of GFP imaging in cancer. University of California, San Diego : AntiCancer, Inc., Dept. of Surgery.
Michiels, N. K., Anthes, N., Hart, N. S., Herler, J., Mexiner, A.J., Schleifenbaum, F., … Wucherer, M. F. (2008). Red fluorescence in reef fish: A novel signalling mechanism? BMC Ecology.
Miyawaki, A. (2005). Innovations in the Imaging of Brain Functions using Fluorescent Proteins. Neuron, 48, 189 – 199.
“National Center for Biotechnology Information.” Current Neurology and Neuroscience Reports., U.S. National Library of Medicine.
Siebeck, U. E., Parker, A. N., Sprenger, D., Mathger, L. M., & Wallis, G. (2010). A Species of Reef Fish that Uses Ultrviolet Patterns for Covert Face Recognition. Current Biology 20, 407-410.
Sparks, J. S., Schelly, R. C., Smith, W. L., Davis, M. P., Tchernov, D., Pieribone, V. A., & Gruber, D. F. (2014). The Covert World of Fish Biofluorescence: A Phylogenetically Widespread and Phenotypically Variable Phenomenon. PLOS ONE journals.
Tsien, R.Y. (1998). The Green Fluorescent Protein. Howard Hughes Medical Institute; University of California, San Diego: Annual Reviews.
BIBLIOGRAPHY
Chudakov, D. M., Matz, M. V., Lukyanov, S., & Lukyanov, K. A. (2010). Fluorescent Proteins and Their Applications in Imaging Living Cells and Tissues. Physiol Rev, 90, 1129 – 1134.
Goodbred, S. & Occhiogrosso, T. (1979). Method for Photographing Small Fish. The Progressive Fish-Culturist, volume 41, NO. 2, pg. 76-77.
Gruber, D. F., Gaffney, J. P., Mehr, S., DeSalle, R., Sparks, J. S., Platisa, J., & Pieribone, V. A. (2015). Adaptive Evolution of Eel Fluorescent Proteins from Fatty Acid Binding Proteins Produces Bright Fluorescence in the Marine Environment. PLOS ONE journals.
Gruber, D. F. & Sparks, J. S. (2015). First Observation of Fluorescence in Marine Turtles. American Museum Novitates.
Gruber, D. F., Loew, E. R., Deheyn, D. D., Akkaynak, D., Gaffney, J. P., Smith, W. L., … Sparks, J. S. (2016). Biofluorescence in Catsharks (Scyliorhinidae): Fundamental Description and Relevance for Elasmobranch Visual Ecology. Scientific Reports.
Holm, E., Mandrak, N.E., & Burridge, M.E. (2009, 2010). The ROM Field Guide To Freshwater Fishes of Ontario. Toronto, Ontario : Royal Ontario Museum.
Wucherer, M. F. & Michiels, N. K. (2012). A Fluorescent Chromatophore Changes the Level of Fluorescence in a Reef Fish. PLOS ONE journals.
SOPHIA COTTRILL
My name is Sophia Cottrill and I am a grade nine student at Owen Sound District Secondary School. I am 15 years old and live in Tara, Ontario. I enjoy studying multiple fields of science, as well as math and music. Outside of school, I participate in a number of extra-curricular activities such as soccer, running, envirothon, and playing saxophone and piano. Science Fair has been a huge part of my life since the time I was eight years old. I have competed at two Canada Wide Science Fairs, earning a gold medal and environmental challenge award in 2016, and a silver medal in 2018. In 2017, I was very fortunate to have been selected as the Canadian Broadcom MASTERS International Delegate, a program that runs alongside the International Science and Engineering Fair. My advice to other students is to do what inspires them and to not be afraid to question the unknown! Science is my passion, and in the future, I plan to pursue a career in biology or medicine.